2.1. Basis Pursuit Denoising via ADMM¶
We are given the measurements \(b = Ax + e\) where \(e\) is a measurement noise.
We solve the problem:
\[
\tag{L1/L2}
{\min}_{x} \| x\|_{1} + \frac{1}{2\rho}\| A x - b \|_2^2
\]
We will work with a sensing matrix \(A\) of size \(M \times N\) which consists of orthonormal rows.
%load_ext autoreload
%autoreload 2
from jax.config import config
config.update("jax_enable_x64", True)
from jax import jit, random
import jax.numpy as jnp
import numpy as np
np.set_printoptions(precision=6)
from jax.numpy.linalg import norm
import matplotlib as mpl
import matplotlib.pyplot as plt
%matplotlib inline
import cr.sparse as crs
import cr.sparse.dict as crdict
import cr.sparse.data as crdata
from cr.sparse import lop
from cr.sparse.cvx.adm import yall1
# Problem size
M = 2000
N = 20000
K = 200
# Dictionary Setup
A = crdict.random_orthonormal_rows(crs.KEYS[0],M, N)
fig=plt.figure(figsize=(8,6), dpi= 100, facecolor='w', edgecolor='k')
plt.imshow(A, extent=[0, 2, 0, 1])
plt.gray()
plt.colorbar()
plt.title(r'$A$');
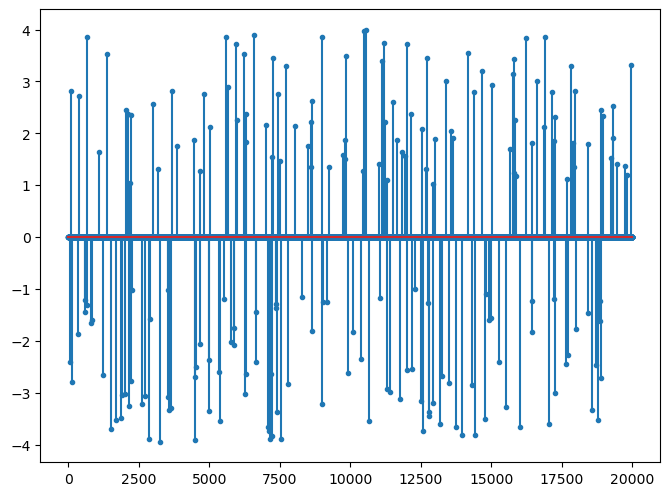
x, omega = crdata.sparse_biuniform_representations(crs.KEYS[1], 1, 4, N, K)
fig=plt.figure(figsize=(8,6), dpi= 100, facecolor='w', edgecolor='k')
plt.stem(x, markerfmt='.');
/tmp/ipykernel_904/1988163945.py:3: UserWarning: In Matplotlib 3.3 individual lines on a stem plot will be added as a LineCollection instead of individual lines. This significantly improves the performance of a stem plot. To remove this warning and switch to the new behaviour, set the "use_line_collection" keyword argument to True.
plt.stem(x, markerfmt='.');
# Convert A into a linear operator
T = lop.real_matrix(A)
T = lop.jit(T)
# Compute the measurements
b0 = T.times(x)
# Generate some Gaussian noise
sigma = 0.01
noise = sigma * random.normal(crs.KEYS[2], (M,))
# Measure the SNR
crs.snr(b0, noise)
DeviceArray(28.29427, dtype=float64)
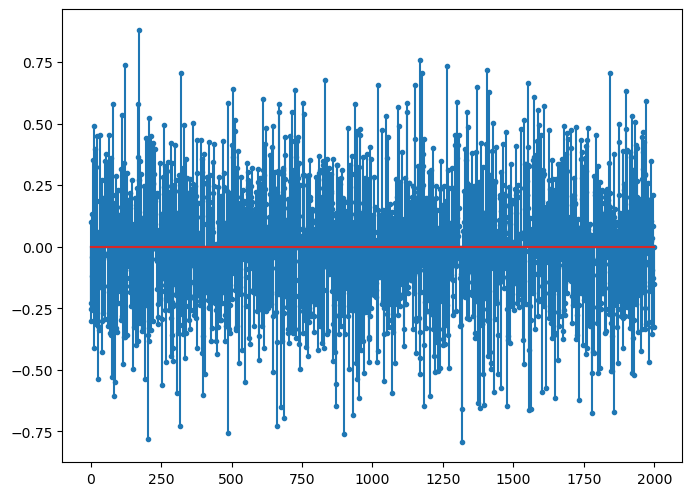
# Add measurement noise
b = b0 + noise
fig=plt.figure(figsize=(8,6), dpi= 100, facecolor='w', edgecolor='k')
plt.stem(b, markerfmt='.');
/tmp/ipykernel_904/1028835729.py:2: UserWarning: In Matplotlib 3.3 individual lines on a stem plot will be added as a LineCollection instead of individual lines. This significantly improves the performance of a stem plot. To remove this warning and switch to the new behaviour, set the "use_line_collection" keyword argument to True.
plt.stem(b, markerfmt='.');
# Solve the BPDN problem
sol = yall1.solve(T, b, rho=sigma)
int(sol.iterations), int(sol.n_times), int(sol.n_trans)
(36, 73, 38)
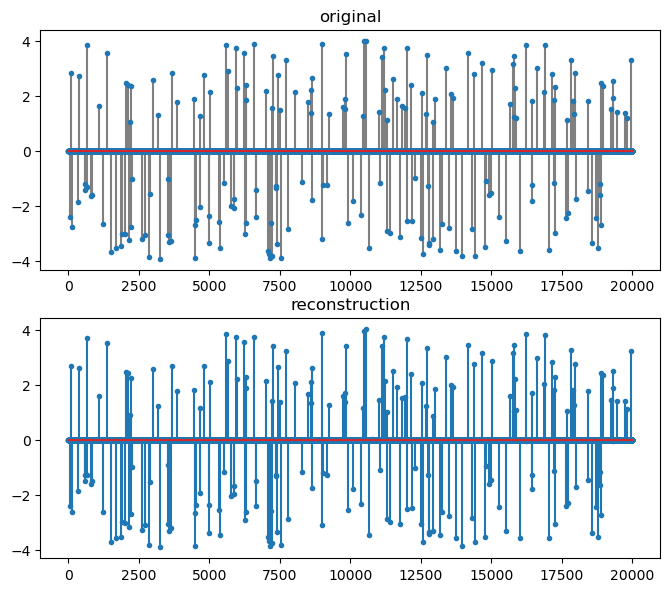
norm(sol.x-x)/norm(x)
DeviceArray(0.026971, dtype=float64)
# The support of K largest non-zero entries in sol.x
omega_rec = crs.largest_indices(sol.x, K)
common = jnp.intersect1d(omega, omega_rec)
total = jnp.union1d(omega, omega_rec)
support_overlap_ratio = len(common) / len(total)
print(support_overlap_ratio)
1.0
fig=plt.figure(figsize=(8,7), dpi= 100, facecolor='w', edgecolor='k')
plt.subplot(211)
plt.title('original')
plt.stem(x, markerfmt='.', linefmt='gray');
plt.subplot(212)
plt.stem(sol.x, markerfmt='.');
plt.title('reconstruction');
/tmp/ipykernel_904/2976149975.py:4: UserWarning: In Matplotlib 3.3 individual lines on a stem plot will be added as a LineCollection instead of individual lines. This significantly improves the performance of a stem plot. To remove this warning and switch to the new behaviour, set the "use_line_collection" keyword argument to True.
plt.stem(x, markerfmt='.', linefmt='gray');
/tmp/ipykernel_904/2976149975.py:6: UserWarning: In Matplotlib 3.3 individual lines on a stem plot will be added as a LineCollection instead of individual lines. This significantly improves the performance of a stem plot. To remove this warning and switch to the new behaviour, set the "use_line_collection" keyword argument to True.
plt.stem(sol.x, markerfmt='.');
%timeit yall1.solve(T, b, rho=sigma).x.block_until_ready()
123 ms ± 45.2 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)
1572 / 273
5.758241758241758